| Copyright | (c) Douglas Burke 2019 |
|---|---|
| License | BSD3 |
| Maintainer | dburke.gw@gmail.com |
| Safe Haskell | None |
| Language | Haskell2010 |
Graphics.Vega.Tutorials.VegaLite
Contents
Description
This tutorial is inspired by - in that it starts off as a close copy of - the
Elm Vega-Lite walkthrough
created by Jo Wood, and
converted as necessary for the differences between hvega and
elm-vegalite.
The Elm tutorial is based on the talk given by
Wongsuphasawat et al at the 2017 Open Vis Conf.
The tutorial targets version 3 of the Vega-Lite specification and
the functionality provided in version 0.4.0.0 of hvega.
Synopsis
- stripPlot :: VegaLite
- stripPlotWithBackground :: VegaLite
- stripPlotY :: VegaLite
- gaiaData :: Data
- stripPlotWithColor :: VegaLite
- stripPlotWithColorOrdinal :: VegaLite
- parallaxBreakdown :: VegaLite
- simpleHistogram :: Text -> VegaLite
- parallaxHistogram :: VegaLite
- gmagHistogram :: VegaLite
- ylogHistogram :: VegaLite
- gmagHistogramWithColor :: VegaLite
- gmagLineWithColor :: VegaLite
- yHistogram :: VegaLite
- pointPlot :: VegaLite
- posPlot :: VegaLite
- skyPlot :: VegaLite
- skyPlot2 :: VegaLite
- smallMultiples :: VegaLite
- smallMultiples2 :: VegaLite
- basePlot :: VegaLite
- layeredPlot :: VegaLite
- layeredDiversion :: VegaLite
- concatenatedPlot :: VegaLite
- concatenatedPlot2 :: VegaLite
- repeatPlot :: VegaLite
- splomPlot :: VegaLite
- selectionProperties :: Text -> Text -> [PropertySpec]
- singleSelection :: VegaLite
- nearestSelection :: VegaLite
- multiSelection :: VegaLite
- eventSelection :: VegaLite
- intervalSelection :: VegaLite
- intervalSelectionY :: VegaLite
- transformSelection :: VegaLite
- widgetSelection :: VegaLite
- bindScales :: VegaLite
- coordinatedViews :: VegaLite
- coordinatedViews2 :: VegaLite
- contextAndFocus :: VegaLite
- crossFilter :: VegaLite
- errorManual :: VegaLite
- errorAuto :: VegaLite
- errorBars :: VegaLite
- errorBand :: VegaLite
- errorBox :: VegaLite
- comparingErrors :: VegaLite
- combinedPlot :: VegaLite
A Grammar of Graphics
hvega is a wrapper for the Vega-Lite visualization grammar which itself is based on Leland Wilkinson's Grammar of Graphics. The grammar provides an expressive way to define how data are represented graphically. The seven key elements of the grammar as represented in hvega and Vega-Lite are:
Data- The input to visualize. Example functions:
dataFromUrl,dataFromColumns, anddataFromRows. Transform- Functions to change the data before they are visualized. Example functions:
filter,calculateAs, andbinAs. These functions are combined withtransform. Projection- The mapping of 3d global geospatial locations onto a 2d plane . Example function:
projection. Mark- The visual symbol, or symbols, that represent the data. Example types, used with
mark:Line,Circle,Bar,Text, andGeoshape. There are also ways to specify the shape to use for thePointtype, using theMShapesetting and theSymboltype. Encoding- The specification of which data elements are mapped to which mark characteristics (commonly known as channels). Example functions:
position,shape,size, andcolor. These encodings are combined withencoding. Scale- Descriptions of the way encoded marks represent the data. Example settings:
SDomain,SPadding, andSInterpolate. Guides- Supplementary visual elements that support interpreting the visualization. Example setings:
AxDomain(for position encodings) andLeTitle(for legend color, size, and shape encodings).
In common with other languages that build upon a grammar of graphics such as D3 and Vega, this grammar allows fine grain control of visualization design. Unlike those languages, Vega-Lite - and hvega in turn - provide practical default specifications for most of the grammar, allowing for a much more compact high-level form of expression.
The Vega-Lite Example Gallery provides a large-number of example visualizations that show off the capabilities of Vega-Lite. Hopefully, by the end of this tutorial, you will be able to create most of them.
How many Haskell extensions do you need?
The VegaLite module is long, but does not use any
complex type machinery, and so it can be loaded without any extensions,
although the extensive use of the Text type means that
using the OverloadedStrings extension is strongly advised.
The module does export several types that conflict with the Prelude, so one suggestion is to use
import Prelude hiding (filter, lookup, repeat)
A note on type safety
The interface provided by hvega provides limited type safety. Various
fields such as PmType are limited by the type of the argument (in this
case Measurement), but there's no support to check that the type makes
sense for the particular column (as hvega itself does not inspect the
data source). Similarly, hvega does not stop you from defining
properties that are not valid for a given situation - for instance
you can say toVegaLite []
Comparing hvega to Elm Vega-Lite
hvega started out as a direct copy of
elm-vegalite,
and has been updated to try and match the functionality of that package.
However, hvega has not (yet?) followed elm-vegalite into using
functions rather than data structures to define the options: for
example, elm-vegalite provides pQuant n which in hvega is the
combination of PName nPmType Quantitativehvega.
The top-level functions - such as dataFromUrl, encoding, and
filter - are generally the same.
What data are we using?
Rather than use the Seattle weather dataset, used in the Elm walkthrough
(if you go through the Vega-Lite Example Gallery
you may also want to look at different data ;-), I am going to use a
small datset from the Gaia satellite,
which has - and still is, as of Summer 2019 - radically-improved our knowledge
of our Galaxy. The data itself is from the paper
"Gaia Data Release 2: Observational Hertzsprung-Russell diagrams"
preprint on arXiV
NASA ADS link.
We are going to use Table 1a, which was downloaded from the
VizieR archive
as a tab-separated file (aka TSV format).
The file contains basic measurements for a number of stars in
nine open clusters that all lie within 250 parsecs of the Earth
(please note, a parsec is a measure of distance, not time, no matter
what some ruggedly-handsome ex-carpenter
might claim). The downloaded file is called
gaia-aa-616-a10-table1a.no-header.tsv, although I have
manually edited it to a "more standard" TSV form (we Astronomers like
our metadata, and tend to stick it in inappropriate places, such as the
start of comma- and tab-separated files, which really mucks up
other-people's parsing code). The first few rows in the file are:
| Source | Cluster | RA_ICRS | DE_ICRS | Gmag | plx | e_plx |
|---|---|---|---|---|---|---|
| 49520255665123328 | Hyades | 064.87461 | +21.75372 | 12.861 | 20.866 | 0.033 |
| 49729231594420096 | Hyades | 060.20378 | +18.19388 | 5.790 | 21.789 | 0.045 |
| 51383893515451392 | Hyades | 059.80696 | +20.42805 | 12.570 | 22.737 | 0.006 |
| ... | ... | ... | ... | ... | ... | ... |
The Source column is a numeric identifier for the star in the Gaia database,
in this particular case the "DR2" release,
the Cluster column tells us which Star Cluster
the star belongs to, RA_ICRS and DE_ICRS
locate the star on the sky
and use the Equatorial coordinate system
(the ICRS term has a meaning too, but it isn't important for our
purposes),
Gmag measues the "brightness" of the star (as in most-things Astronomical,
this is not as obvious as you might think, as I'll go into below),
and the plx and e_plx columns give the measured
parallax of the star
and its error value, in units of
milli arcseconds.
And yes, I do realise after complaining about popular-culture references
confusing distances and time, I am now measuring distances with angles.
I think I've already mentioned that Astronomy is confusing...
Creating the Vega-Lite visualization
The function toVegaLite takes a list of grammar specifications,
as will be shown in the examples below, and creates a single JSON object
that encodes the entire design. As of hvega-0.4.0.0 this targets
version 3 of the Vega-Lite schema, but this can be over-ridden with
toVegaLiteSchema if needed.
There is no concept of ordering to these specification lists, in that
[ dataFromUrl ..., encoding ..., mark ...];
[ encoding ..., dataFromUrl ..., mark ... ];
and
[ encoding ..., mark ..., dataFromUrl ... ]
would all result in the same visualization.
The output of toVegaLite can be sent to the Vega-Lite runtime to
generate the Canvas or SVG output. hvega contains the helper
routines:
fromVL, which is used to extract the JSON contents fromVegaLiteand create an AesonValue;toHtml, which creates a HTML page which uses the Vega Embed Javascript library to display the Vega-Lite visualization;- and
toHtmlFile, which is liketoHtmlbut writes the output to a file.
A Strip Plot
In this section we shall concentrate on creating a single plot. Later on we shall try combining plots, after branching out to explore some of the different ways to visualize multi-dimensional data sets.
In the examples I link to symbols that have not been used in previous visualizations, to make it easier to see the use of new functionality.
Our first hvega plot
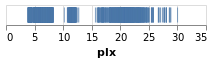
stripPlot :: VegaLite Source #
We could encode one of the numeric data fields as a strip plot where
the horizontal position of a tick mark is determined by the value
of the data item. In this case I am going to pick the "plx" column:
Open this visualization in the Vega Editor
toVegaLite[dataFromUrl"https://raw.githubusercontent.com/DougBurke/hvega/master/hvega/data/gaia-aa-616-a10-table1a.no-header.tsv" [TSV] ,markTick[] ,encoding(positionX[PName"plx",PmTypeQuantitative] []) ]
Notice how there is no explicit definition of the axis details, color choice or size. These can be customised, as shown in examples below, but the default values are designed to follow good practice in visualization design.
Three grammar elements are represented by the three functions
dataFromUrl, mark, and encoding.
The encoding function takes as a single parameter, a list of
specifications that are themselves generated by other functions. In
this case we use the function position to provide an encoding of the
"plx" field as the x-position in our plot. The precise way in which
the data value (parallax) is mapped to the x-position will depend on the type of
data we are encoding. We can provide a hint by delcaring the
measurement type of the data field, here Quantitative indicating a
numeric measurement type. The final parameter of position is a list of
any additional encodings in our specification. Here, with only one
encoding, we provide an empty list.
As we build up more complex visualizations we will use many more encodings. To keep the coding clear, the idiomatic way to do this with hvega is to chain encoding functions using point-free style. The example above coded in this way would be
let enc = encoding
. position X [ PName "plx", PmType Quantitative ]
in toVegaLite
[ dataFromUrl "https://raw.githubusercontent.com/DougBurke/hvega/master/hvega/data/gaia-aa-616-a10-table1a.no-header.tsv" [TSV]
, mark Tick []
, enc []
]
Backgrounds
The default background color for the visualization, at least in the
Vega-Embed PNG and SVG output, is transparent. In many cases this is
perfectly fine, but an explicit color can be specified using the
Background configuration option.
stripPlotWithBackground :: VegaLite Source #
The configure function allows a large number of configuration
options to be configured, each one introduced by the
configuration function. Here I set the color to be a light gray
(actually a very-transparent black; the Color type describes the
various supported color specifications, but it is generally safe to assume
that if you can use it in HTML then you can use it here).
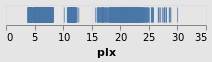
Open this visualization in the Vega Editor
let enc = encoding
. position X [ PName "plx", PmType Quantitative ]
conf = configure
. configuration (Background "rgba(0, 0, 0, 0.1)")
in toVegaLite
[ dataFromUrl "https://raw.githubusercontent.com/DougBurke/hvega/master/hvega/data/gaia-aa-616-a10-table1a.no-header.tsv" [TSV]
, mark Tick []
, enc []
, conf []
]
Challenging the primacy of the x axis
stripPlotY :: VegaLite Source #
There is nothing that forces us to use the x axis, so let's
try a vertical strip plot. To do so requires changing only
one character in the specifiction, that is the first argument to
position is now Y rather than X:
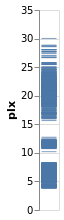
Open this visualization in the Vega Editor
let enc = encoding
. position Y [ PName "plx", PmType Quantitative ]
in toVegaLite
[ dataFromUrl "https://raw.githubusercontent.com/DougBurke/hvega/master/hvega/data/gaia-aa-616-a10-table1a.no-header.tsv" [TSV]
, mark Tick []
, enc []
]
Data sources
Since we are going to be using the same data source, let's define it here:
gaiaData = let addFormat n = (n,FoNumber) cols = [ "RA_ICRS", "DE_ICRS", "Gmag", "plx", "e_plx" ] opts = [Parse(map addFormat cols) ] in dataFromUrl "https://raw.githubusercontent.com/DougBurke/hvega/master/hvega/data/gaia-aa-616-a10-table1a.no-header.tsv" opts
The list argument to dataFromUrl allows for some customisation of
the input data. Previously I used [ to specify the data is in
tab-separated format, but it isn't actually needed here (since the
file name ends in ".tsv"). However, I have now explicitly defined how
to parse the numeric columns using TSV]Parse: this is because the columns
are read in as strings for this file by default, which actually doesn't
cause any problems in most cases, but did cause me significant problems
at one point during the development of the tutorial! There is limited
to no feedback from the visualizer for cases like this (perhaps I should
have used the Javascript console), and I only realised the problem thanks
to the Data Viewer tab in the
Vega Editor
(after a
suggestion from a colleague).
Although not used in this tutorial, data
can also be defined algorithmically - using dataSequence and
dataSequenceAs - or inline - with dataFromColumns or
dataFromRows - or directly from JSON (as a Value) using
dataFromJson.
Adding color as an encoding
stripPlotWithColor :: VegaLite Source #
One question would be how the parallaxes vary by cluster: as parallax is measuring distance, then are the clusters similar distances away from us, or are there a range of values? A first look is to use another "channel" to represent (i.e. encode) the cluster:
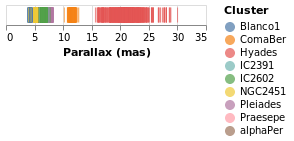
Open this visualization in the Vega Editor
let enc = encoding
. position X [ PName "plx", PmType Quantitative, PAxis [ AxTitle "Parallax (mas)" ] ]
. color [ MName "Cluster", MmType Nominal ]
in toVegaLite
[ gaiaData
, mark Tick []
, enc []
]
Now each tick mark is colored by the cluster, and a legend is automatically
added to indicate this mapping. Fortunately the number of clusters in the
sample is small enough to make this readable! The color function has
added this mapping, just by giving the column to use (with MName) and
its type (MmType). The constructors generally begin with P for
position and M for mark, and as we'll see there are other property
types such as facet and text.
Vega-Lite supports several data types, represented
by the Measurement type. We have already seen Quantitative, which
is used for numeric data, and here we use Nominal for the clusters,
since they have no obvious ordering.
The labelling for the X axis has been tweaked using PAxis, in this
case the default value for the label (the column name) has been
over-ridden by an explicit value.
Comparing Ordinal with Nominal data types
It is instructive to see what happens if we change the mark type for
the color encoding from Nominal to Ordinal.
stripPlotWithColorOrdinal :: VegaLite Source #

Open this visualization in the Vega Editor
let enc = encoding
. position X [ PName "plx", PmType Quantitative, PAxis [ AxTitle "Parallax (mas)" ] ]
. color [ MName "Cluster", MmType Ordinal ]
in toVegaLite
[ gaiaData
, mark Tick []
, enc []
]
As can be seen, the choice of color scale has changed to one more appropriate for an ordered set of values.
Adding an axis
While the strip plot shows the range of parallaxes, it is hard to
make out the distribution of values, since the ticks overlap. Even
changing the opacity of the ticks - by adding an encoding channel
like opacity [ MNumber 0.6 ]MOpacity
property of the mark - only helps so much. Adding a second
axis is easy to do, so let's see how the parallax distribution
varies with cluster membership.
parallaxBreakdown :: VegaLite Source #
The stripPlotWithColor visualization can be changed to show two
variables just by adding a second position declaration:
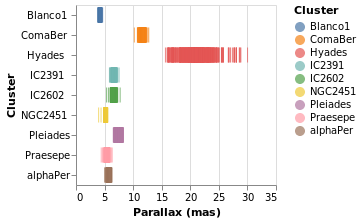
Open this visualization in the Vega Editor
let enc = encoding
. position X [ PName "plx", PmType Quantitative, PAxis [ AxTitle "Parallax (mas)" ] ]
. position Y [ PName "Cluster", PmType Nominal ]
. color [ MName "Cluster", MmType Nominal ]
in toVegaLite
[ gaiaData
, mark Tick []
, enc []
]
I have left the color-encoding in, as it makes it easier to compare to
stripPlotWithColor, even though it replicates the information provided
by the position of the mark on the Y axis.
Creating a value to plot: aggregating data
We can also "create" data to be plotted, by aggregating data. In this case we can create a histogram showing the number of stars with the same parallax value (well, a range of parallaxes).
simpleHistogram :: Text -> VegaLite Source #
Since sensible (hopefully) defaults are provided for unspecified settings, it is relatively easy to write generic representations of a particular visualization. The following function expands upon the previous specifications by:
- taking a field name, rather than hard coding it;
- the use of
PBin[] - the addition of a second axis (
Y) which is used for the aggregated value (Count, which means that no column has to be specified withPName); - and the change from
TicktoBarfor themark.
Note that we did not have to specify how we wanted the histogram
calculation to proceed - e.g. the number of bins, the bin widths,
or edges - although we could have added this, by using a non-empty
list of BinProperty values with PBin, if the defaults are not
sufficient.
simpleHistogram field =
let enc = encoding
. position X [ PName field, PmType Quantitative, PBin [] ]
. position Y [ PAggregate Count, PmType Quantitative ]
in toVegaLite
[ gaiaData
, mark Bar []
, enc []
]
parallaxHistogram :: VegaLite Source #
With simpleHistogram it becomes easy to get a histogram of the parallax
values:
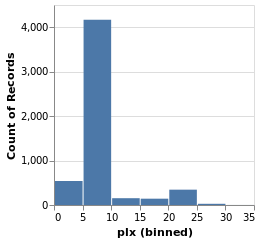
Open this visualization in the Vega Editor
parallaxHistogram = simpleHistogram "plx"We can see that although parallaxes around 20 to 25 milli-arcseconds
dominated the earlier visualizations, such as stripPlotWithColor,
most of the stars have a much-smalled parallax, with values
in the range 5 to 10.
gmagHistogram :: VegaLite Source #
A different column (or field) of the input data can be viewed, just by changing the name in the specification:

Open this visualization in the Vega Editor
gmagHistogram = simpleHistogram "Gmag"
Here we can see that the number of stars with a given magnitude rises up until a value of around 18, and then drops off.
Changing the scale of an axis
In the case of parallaxHistogram, the data is dominated by
stars with small parallaxes. Changing the scale of the
Y axis to use a logarithmic, rather than linear, scale might
provide more information:
ylogHistogram :: VegaLite Source #
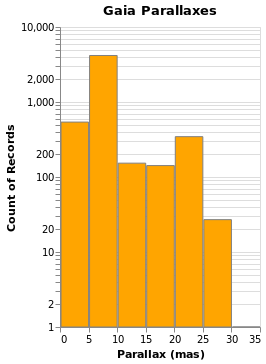
Open this visualization in the Vega Editor
let enc = encoding
. position X [ PName "plx", PmType Quantitative, PBin [], PAxis [ AxTitle "Parallax (mas)" ] ]
. position Y [ PAggregate Count, PmType Quantitative, PScale [ SType ScLog ] ]
in toVegaLite
[ gaiaData
, mark Bar [ MFill "orange", MStroke "gray" ]
, enc []
, height 300
, title "Gaia Parallaxes" []
]
There are four new changes to the visualization created by simpleHistogram (since PAxis
has been used above):
- an explicit choice of scaling for the Y channel (using
PScale); - the fill (
MFill) and edge (MStroke) colors of the histogram bars are different; - the height of the overall visualization has been increased;
- and a title has been added.
If you view this in the Vega Editor you will see the following warning:
A log scale is used to encode bar's y. This can be misleading as the height of the bar can be arbitrary based on the scale domain. You may want to use point mark instead.
Stacked Histogram
gmagHistogramWithColor :: VegaLite Source #
A color encoding can also be added. When used with the Tick mark -
stripPlotWithColor - the result was that each tick mark was colored
by the "Cluster" field, but for the Bar mark the result is that
the bars are stacked together. I have also taken the opportunity to
widen the plot (width); define the binning scheme used, with Step
1AxValues.
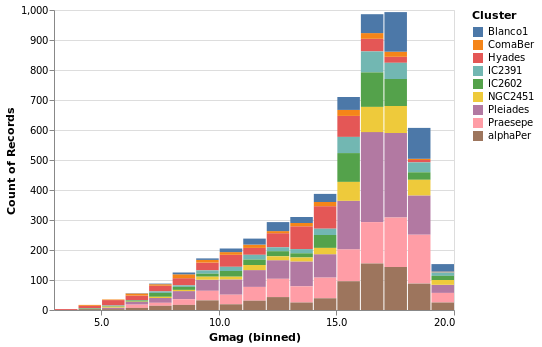
Open this visualization in the Vega Editor
let enc = encoding
. position X [ PName "Gmag", PmType Quantitative, binning, axis ]
. position Y [ PAggregate Count, PmType Quantitative ]
. color [ MName "Cluster", MmType Nominal ]
binning = PBin [ Step 1 ]
axis = PAxis [ AxValues (Numbers [ 0, 5 .. 20 ]) ]
in toVegaLite
[ gaiaData
, mark Bar []
, enc []
, height 300
, width 400
]
Note that hvega will allow you to combine a color encoding with a ScLog
scale, even though a Vega-Lite viewer will not display the
resulting Vega-Lite specification, saying
Cannot stack non-linear scale (log)
gmagLineWithColor :: VegaLite Source #
Notice how we never needed to state explicitly that we wished our bars
to be stacked. This was reasoned directly by Vega-Lite based on the
combination of bar marks and color channel encoding. If we were to
change just the mark function from Bar to Line, Vega-Lite produces an
unstacked series of lines, which makes sense because unlike bars,
lines do not occlude one another to the same extent.
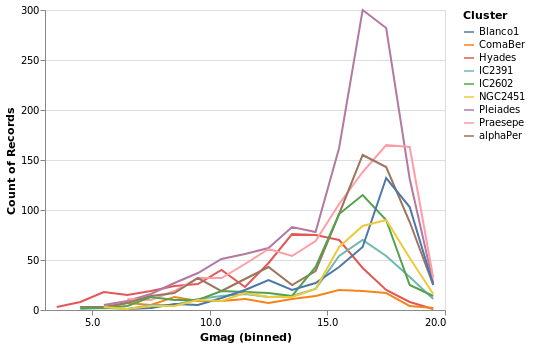
Open this visualization in the Vega Editor
let enc = encoding
. position X [ PName "Gmag", PmType Quantitative, binning, axis ]
. position Y [ PAggregate Count, PmType Quantitative ]
. color [ MName "Cluster", MmType Nominal ]
binning = PBin [ Step 1 ]
axis = PAxis [ AxValues (Numbers [ 0, 5 .. 20 ]) ]
in toVegaLite
[ gaiaData
, mark Line []
, enc []
, height 300
, width 400
]
You don't have to just count
The previous histogram visualizations have taken advantage of Vega-Lite's
ability to bin up (Count) a field, but there are a number of aggregation
properties (as defined by the Operation type). For example, there
are a number of measures of the "spread" of a population, such as
the sample standard deviation (Stdev).
yHistogram :: VegaLite Source #
The aim for this visualization is to show the spread in the Gmag field
for each cluster, so we now swap the axis on which the aggregate is
being applied (so that the cluster names appear on the y axis),
and hide the legend that is applied (using MLegend []
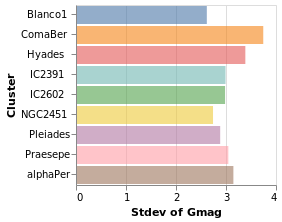
Open this visualization in the Vega Editor
let enc = encoding
. position X [ PName "Gmag", PmType Quantitative, PAggregate Stdev ]
. position Y [ PName "Cluster", PmType Nominal ]
. color [ MName "Cluster", MmType Nominal, MLegend [] ]
in toVegaLite
[ gaiaData
, mark Bar [ MOpacity 0.6 ]
, enc []
]
The bar opacity is reduced slightly with 'MOpacity 0.6' so that the
x-axis grid lines are visible. An alternative would be to change the
AxZIndex value for the X encoding so that it is drawn on top of
the bars.
Plotting with points
At this point we make a signifiant detour from the Elm Vega-Lite
walkthtough, and look at the Point mark, rather than creating
small-multiple plots. Don't worry, we'll get to them later.
pointPlot :: VegaLite Source #
Here I use the Point mark to display the individual
Gmag, plx pairs, encoding by both color and 'shape.
Since the encoding uses the same field of the data (the Cluster
name), Vega-Lite is smart enough to only display one legend,
which contains the point shape and color used for each cluster.
Since the parallax values are bunched together at low values,
a logarithmic scale (ScLog) is used for the y axis, along with
commands to define the actual axis domain - by turning off the
IsNice support and listing the minimum and maximum values
for the axis with SDomain.
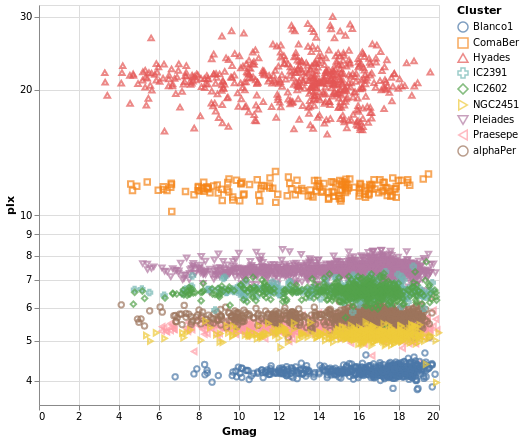
Open this visualization in the Vega Editor
let enc = encoding
. position X [ PName "Gmag", PmType Quantitative ]
. position Y [ PName "plx", PmType Quantitative, PScale scaleOpts ]
. color cluster
. shape cluster
scaleOpts = [ SType ScLog, SDomain (DNumbers [3.5, 32]), SNice (IsNice False) ]
cluster = [ MName "Cluster", MmType Nominal ]
in toVegaLite [ gaiaData
, mark Point []
, enc []
, width 400
, height 400
]
We can see that each cluster appears to have a separate parallax value, and that it doesn't really vary with Gmag. What this is telling us is that for these star clusters, the distance to each member star is similar, and that they are generally at different distances from us. However, it's a bit hard to tell exactly what is going on around 5 to 6 milli arcseconds, as the clusters overlap here.
This line of thinking leads us nicely to map making, but before we
try some cartography, I wanted to briefly provide some context for
these plots. The
parallax of a star
is a measure of its distance from us, but it is an inverse relationship,
so that nearer stars have a larger parallax than those further from us.
The Gmag column measures the apparent brightness of the star, with the
G part indicating what
part of the spectrum
is used (for Gaia, the G band is pretty broad, covering much of
the visible spectrum), and the mag part is because optical Astronomy
tends to use
- the logarithm of the measured flux
- and then subtract this from a constant
so that larger values mean fainter sources. These are also apparent magnitues, so that they measure the flux of the star as measured at Earth, rather than its intrinsic luminosity (often defined as an object's absolute magnitude).
We can see that the further the cluster is from us - that is, as we move down this graph to smaller parallax values - then the smallest stellar magnitude we can see in a cluster tends to increase, but that there are stars up to the maximum value (20) in each cluster. This information can be used to look at the distribution of absolute magnitudes of stars in a cluster, which tells us about its evolutionary state - such as is it newly formed or old - amongst other things. However, this is straying far from the aim of this tutorial, so lets get back to plotting things.
Making a map
We have some hint that the different clusters are distinct objects
in space, in that they appear to be different distances from us,
but where does the "cluster" in the name "Stellar Cluster"
come from? Well, we can try plotting up the position of each star
on the sky - using the RA_ICRS and DE_ICRS fields - to find out.
The following specification should only contain one new feature - other
than sneakily switching from Point to Circle type for the mark - and
that is displaying the x axis (namely Right Ascension) in reverse (using
PSort [ Descending ]
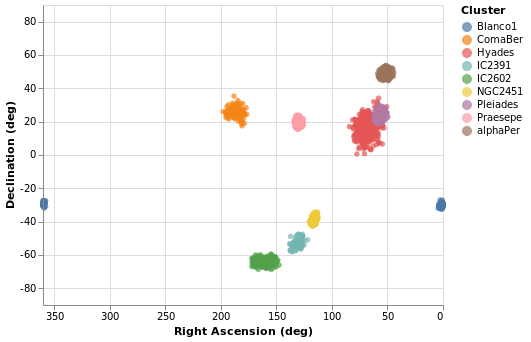
Open this visualization in the Vega Editor
let enc = encoding
. position X (axOpts "RA_ICRS" "Right Ascension (deg)" ++ [ raScale, PSort [ Descending ] ])
. position Y (axOpts "DE_ICRS" "Declination (deg)" ++ [ decScale ])
. color [ MName "Cluster", MmType Nominal ]
axOpts field lbl = [ PName field, PmType Quantitative, PAxis [ AxTitle lbl ]]
scaleOpts minVal maxVal = [ SDomain (DNumbers [ minVal, maxVal ]), SNice (IsNice False) ]
raScale = PScale (scaleOpts 0 360)
decScale = PScale (scaleOpts (-90) 90)
in toVegaLite [ gaiaData
, mark Circle []
, enc []
, width 400
, height 300
]
We can see that these clusters are indeed localised on the sky, with Hyades looking like it covers the largest area. However, we should be careful and not forget either Grover's hard work or Father Ted's explanation to Father Dougal, since these clusters are different distances from us, which makes size a tricky thing to measure from this plot.
There is also the fact that I have used possibly the worst way of displaying the Right Ascension and Declination data. Although the night sky is not the same as the Earth's surface, the issues when trying to display the Globe on a flat surface also apply to displaying up the sky. For this plot the distortions near the pole are huge, although fortunately we don't have any clusters too close to either pole.
Vega-Lite supports a large number of projections - via the
Projection type - which we use below to create
a similar visualization to posPlot. Here I use the
Longitude and Latitude channels, along with a
Mercator projection, to display the data.
The trick in this case is that longitude runs from -180 to 180
degrees, but the data has Right Ascension going from 0
to 360 degrees. We can take advantage of Vega Lite's
data transformation capabilities and create a new
column - which I call fakeLongitude - and is
defined as "180 - Right Ascension". The "expression" support
is essentially a sub-set of Javascript, and the datum
object refers to the current row. The new data column
can then be used with the Longitude channel.
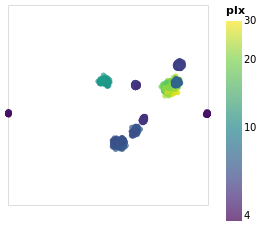
Open this visualization in the Vega Editor
let enc = encoding
. position Longitude (axOpts "fakeLongitude")
. position Latitude (axOpts "DE_ICRS")
. color [ MName "plx"
, MmType Quantitative
, MScale [ SType ScLog
, SScheme "viridis" []
]
]
. tooltip [ TName "Cluster", TmType Nominal ]
axOpts field = [ PName field, PmType Quantitative ]
trans = transform
. calculateAs "180 - datum.RA_ICRS" "fakeLongitude"
in toVegaLite [ projection [ PrType Mercator ]
, gaiaData
, trans []
, mark Circle []
, enc []
]
The other major change made to posPlot is that the stars are now
color-encoded by the log of their parallax value
rather than cluster membership,
and the color scheme has been changed to use the "viridis" color
scale.
Since parallax is a numeric value, with ordering (i.e. Quantitative),
the legend has changed from a list of symbols to a gradient bar.
To account for this lost of information, I have added a tooltip
encoding so that when the pointer is moved over a star its cluster
name will be displayed (this replaces the default behavior when
all the encoded channels, so in this case the position and parallax
values, would be displayed, and it is only visible in "interactive"
versions of the visualization).
From this visualization we can see that the apparent size of the cluster
(if we approximate each cluster as a circle, then we can think of the radius
of the circle as a measure of size) depends on parallax, with larger
sizes having larger parallaxes. This is because the distance to a star
is inversely-dependent on its parallax, so larger parallaxes mean the
star is closer to us. However, there is no reason that the intrinsic
size - that is its actual radius - of each cluster is the same.
We can see that although the Hyades and Pleiades clusters overlap
on the sky, they have significantly-different parallaxes (as can
be seen in pointPlot for example), with Hyades being the closer
of the two.
It is possible to add graticules - with the aptly-named
graticule function - but this appears to need to be done
as a layer, which we haven't covered yet. I may get to it
below, but am having teensy problems with getting it
working in the Vega Lite viewer I use (the one provided with
Jupyter notebook, which defaults to an earlier version
of Vega-Lite support, which doesn't seem to like the more-recent
capabilities of Vega Lite).
Layered and Multi-View Compositions
The Stacked-Histogram plot - created by gmagHistogramWithColor - showed
the distribution of the "Gmag" field by cluster, but it was hard to
compare them. A common approach in this situation is to split up
the data into multiple plots -
the small multiple
approach (also known as trellis plots) - which we can easily achieve in
Vega Lite. It also gets us back on track with the Elm walkthrough.
smallMultiples :: VegaLite Source #
Our first attempt is with the column function, which tells
Vega-Lite to create a plot for each Cluster field (and introduces
us to the F family of FacetChannel constructors).
The legend has been turned off with MLegend []
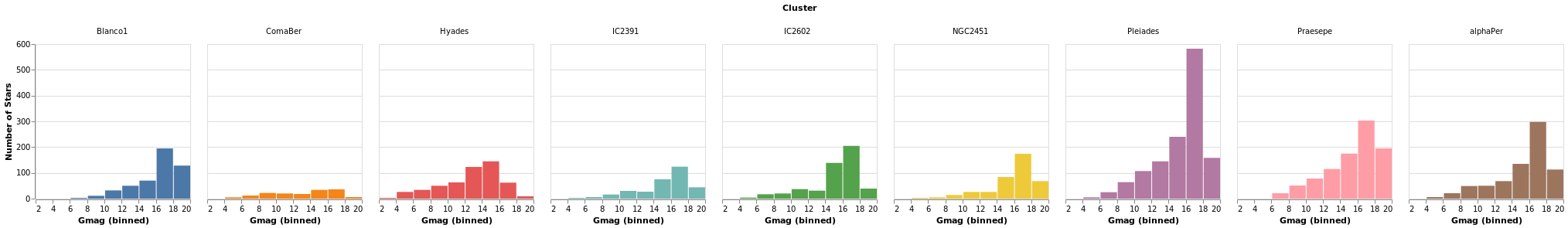
Open this visualization in the Vega Editor
let enc = encoding
. position X [ PName "Gmag", PmType Quantitative, PBin [] ]
. position Y yAxis
. color [ MName "Cluster", MmType Nominal, MLegend [] ]
. column [ FName "Cluster", FmType Nominal ]
yAxis = [ PAggregate Count
, PmType Quantitative
, PAxis [ AxTitle "Number of Stars" ]
]
in toVegaLite
[ gaiaData
, mark Bar []
, enc []
]
Since we have nine clusters in the sample, the overall visualization is too wide, unless you have a very-large monitor. Can we do better?
smallMultiples2 :: VegaLite Source #
The number of columns used in small-multiple can be defined using the
columns function. However, this requires us to:
- move the facet definition out from the encoding and into the top-level,
with the
facetFlowfunction; - and define the plot as a separate specification, and apply it
with
specificationandasSpec.
The actual syntactic changes to smallMultiples are actually
fairly minor:
let enc = encoding
. position X [ PName "Gmag", PmType Quantitative, PBin [] ]
. position Y yAxis
. color [ MName "Cluster", MmType Nominal, MLegend [] ]
yAxis = [ PAggregate Count
, PmType Quantitative
, PAxis [ AxTitle "Number of Stars" ]
]
in toVegaLite
[ gaiaData
, columns 4
, facetFlow [ FName "Cluster", FmType Nominal ]
, specification (asSpec [ mark Bar [], enc [] ])
]
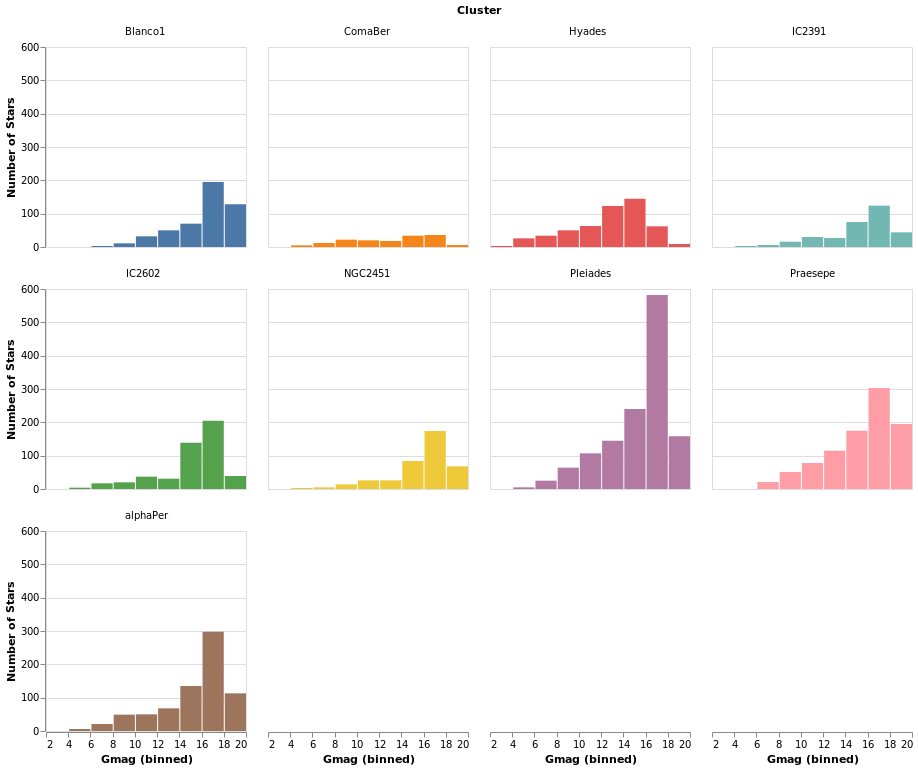
Open this visualization in the Vega Editor
Note that Vega Lite does support a "facet" field in its encodings,
but hvega follows Elm VegaLite and requires you to use this
wrapped facet approach.
One plot, two plot, red plot, blue plot
There are four ways in which multiple views may be combined:
- The facet operator takes subsets of a dataset (facets) and
separately applies the same view specification to each of
those facets (as seen with the
columnfunction above). Available functions to create faceted views:column,row,facet,facetFlow, andspecification. - The layer operator creates different views of the data but
each is layered (superposed) on the same same space; for example
a trend line layered on top of a scatterplot.
Available functions to create a layered view:
layerandasSpec. - The concatenation operator allows arbitrary views (potentially
with different datasets) to be assembled in rows or columns.
This allows 'dashboards' to be built.
Available functions to create concatenated views:
vConcat,hConcat, andasSpec. - The repeat operator is a concise way of combining multiple views
with only small data-driven differences in each view.
Available functions for repeated views:
repeatandspecification.
We start with a "basic" plot for the dataset: the median value of the parallax of the stars in each cluster.
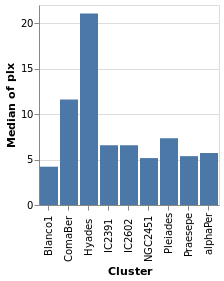
Open this visualization in the Vega Editor
let plx = position Y [ PName "plx", PmType Quantitative, PAggregate Median ]
cluster = position X [ PName "Cluster", PmType Nominal ]
enc = encoding . cluster . plx
in toVegaLite
[ gaiaData
, mark Bar []
, enc []
]
Composing layers
layeredPlot :: VegaLite Source #
We start our exploration by combining two visualizations, layering
one on top of the other. The base plot shows the same data as
basePlot, and then on top we will show a horizontal line that
indicates the median parallax for all the stars in the sample.
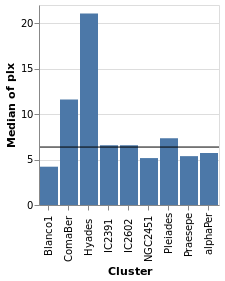
Open this visualization in the Vega Editor
let plx = position Y [ PName "plx", PmType Quantitative, PAggregate Median ]
cluster = position X [ PName "Cluster", PmType Nominal ]
perCluster = [ mark Bar [], encoding (cluster []) ]
allClusters = [ mark Rule [] ]
in toVegaLite
[ gaiaData
, encoding (plx [])
, layer (map asSpec [perCluster, allClusters])
]
For this visualization, the specification starts with the data
source and an encoding, but only for the y axis (which means
that all layered plots use the same encoding for the axis). The
layer function introduces the different visualizations that
will be combined, each as there own "specification" (hence
the need to apply asSpec to both perCluster and allClusters).
Note that there is no x-axis encoding for the Rule, since the
data applies to all clusters (i.e. it should span the
whole visualization).
layeredDiversion :: VegaLite Source #
This example is similar to layeredPlot but includes an x-axis
encoding for the second layer. We use this to show the range of the
data - so the minimum to maximum parallax range of each cluster - with
the Rule type. The difference to the previous plot is that an
extra positional encoding is added (Y2) to define the end point
of each line (Y is used as the start point).
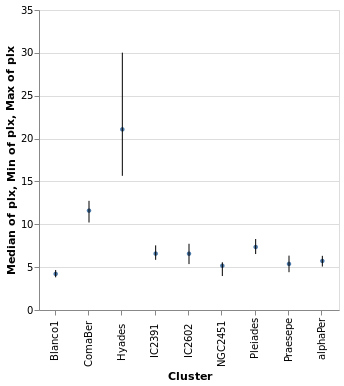
Open this visualization in the Vega Editor
let plx op = position Y [ PName "plx", PmType Quantitative, PAggregate op ]
cluster = position X [ PName "Cluster", PmType Nominal ]
median = [ mark Circle [ MSize 20 ]
, encoding (plx Median [])
]
range = [ mark Rule [ ]
, encoding
. plx Min
. position Y2 [ PName "plx", PAggregate Max ]
$ []
]
in toVegaLite
[ gaiaData
, encoding (cluster [])
, layer (map asSpec [ median, range ])
, width 300
, height 300
]
The MSize option is used to change the size of the circles so that they
do not drown out the lines (the size value indicates the area of the mark,
and so for circles the radius is proportional to the square root of this
size value; in practical terms I adjusted the value until I got something
that looked sensible).
Note that the y axis is automatically labelled with the different operation types that were applied - median, minimum, and maximum - although there is no indication of what marks map to these operations.
Concatenating views
concatenatedPlot :: VegaLite Source #
Instead of layering one view on top of another (superposition), we can place them side by side in a row or column (juxtaposition). In Vega-Lite this is referred to as concatenation:
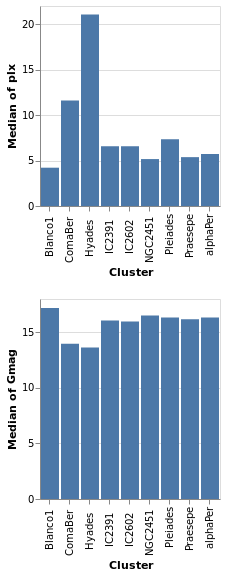
Open this visualization in the Vega Editor
let enc field = encoding
. position X [ PName "Cluster", PmType Nominal ]
. position Y [ PName field, PmType Quantitative, PAggregate Median ]
parallaxes = [ mark Bar [], enc "plx" [] ]
magnitudes = [ mark Bar [], enc "Gmag" [] ]
specs = map asSpec [ parallaxes, magnitudes ]
in toVegaLite
[ gaiaData
, vConcat specs
]
The hConcat function would align the two plots horizontally,
rather than vertically (and is used in combinedPlot).
Note that as the axes are identical apart from the field for the y axis, the encoding has been moved into a function to enforce this constraint (this ensures the x axis is the same, which makes it easier to visually compare the two plots). However, there is no requirement that the two plots be "compatible" (they could use different data sources).
concatenatedPlot2 :: VegaLite Source #
The alignment of the plots can be adjusted with spacing, which we
use here to remove the vertical gap between the two plots (the
example is written so that we can see the only difference between
the two plot specifications is the addition of PAxis []
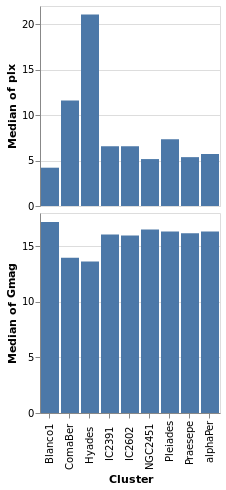
Open this visualization in the Vega Editor
let enc field flag = encoding
. position X ([ PName "Cluster", PmType Nominal ] ++
if flag then [ PAxis [] ] else [])
. position Y [ PName field, PmType Quantitative, PAggregate Median ]
parallaxes = [ mark Bar [], enc "plx" True [] ]
magnitudes = [ mark Bar [], enc "Gmag" False [] ]
specs = map asSpec [ parallaxes, magnitudes ]
in toVegaLite
[ gaiaData
, spacing 0
, vConcat specs
]
Even though we set spacing to 0 there is still a small gap between
the plots: this can be removed by using bounds Flush
Repeated views
Creating the same plot but with a different field is common-enough
that Vega-Lite provides the repeat operator.
repeatPlot :: VegaLite Source #
The concatenatedPlot example can be extended to view the
distribution of several fields - in this case Right Ascension,
Declination, parallax, and magnitude:

Open this visualization in the Vega Editor
let enc = encoding
. position X [ PName "Cluster", PmType Nominal ]
. position Y [ PRepeat Row, PmType Quantitative, PAggregate Median ]
spec = asSpec [ gaiaData
, mark Bar []
, enc [] ]
rows = [ "RA_ICRS", "DE_ICRS", "plx", "Gmag" ]
in toVegaLite
[ repeat [ RowFields rows ]
, specification spec
]
This more compact specification replaces the data field name
(for example PName "plx"PRepeat) either as a Row or Column depending on the desired
layout. We then compose the specifications by providing a set of
RowFields (or ColumnFields) containing a list of the fields to which
we wish to apply the specification (identified with the function
specification which should follow the repeat function provided to
toVegaLite).
splomPlot :: VegaLite Source #
We can combine repeated rows and columns to create a grid of views, such as a scatterplot matrix, adding in color encoding to separate out the clusters:
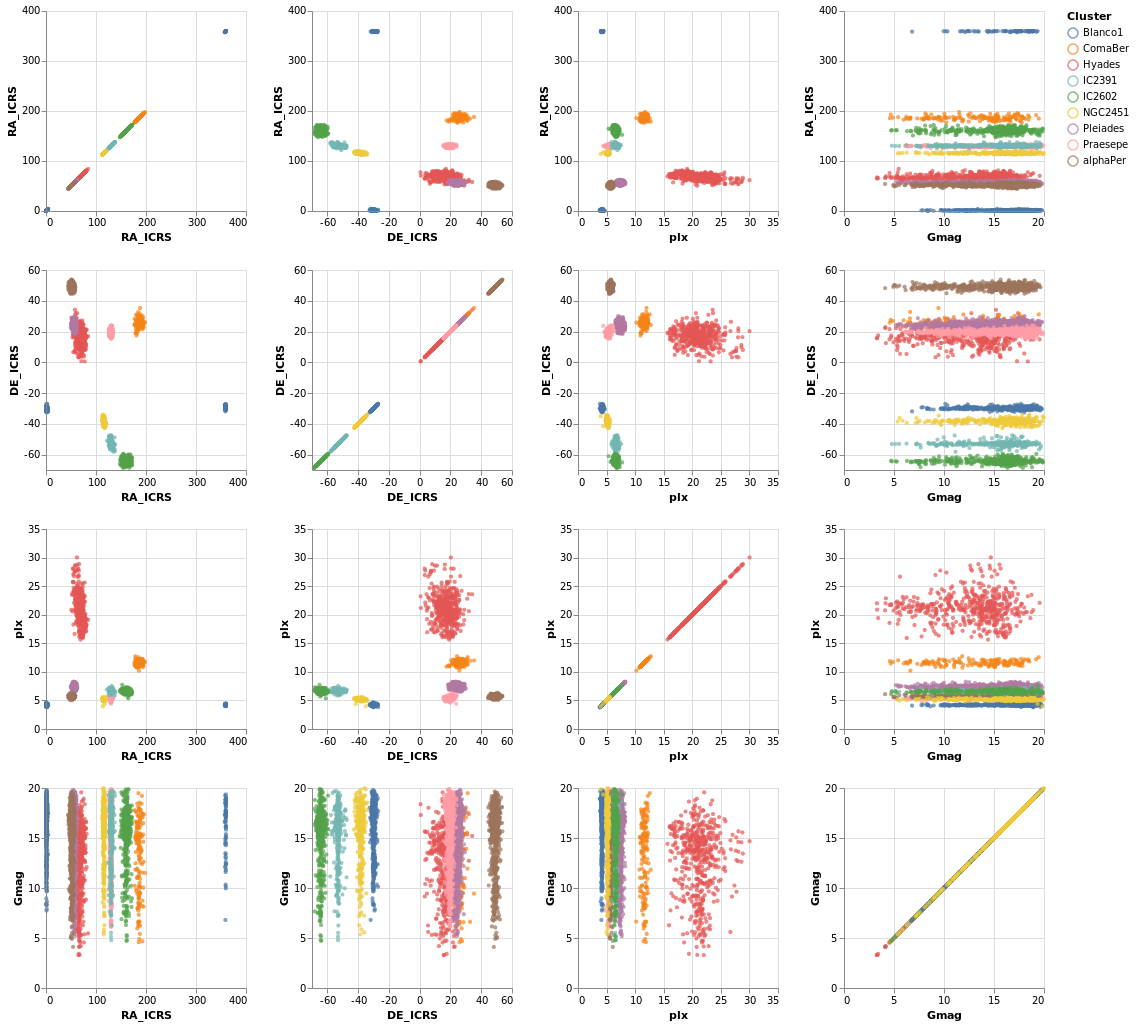
Open this visualization in the Vega Editor
let enc = encoding
. position X [ PRepeat Column, PmType Quantitative ]
. position Y [ PRepeat Row, PmType Quantitative ]
. color [ MName "Cluster", MmType Nominal ]
spec = asSpec [ gaiaData
, mark Point [ MSize 5 ]
, enc [] ]
fields = [ "RA_ICRS", "DE_ICRS", "plx", "Gmag" ]
in toVegaLite
[ repeat [ RowFields fields, ColumnFields fields ]
, specification spec
]
To be honest, this is not the best dataset to use here, as
there is no direct correlation between location (the RA_ICRS
and DE_ICRS fields) and the other columns, but it's the
dataset I chose, so we are stuck with it.
Once you have sub-plots as a specification, you can combine them horizontally and vertically to make a dashboard style visualization. Interested parties should check out the Building a Dashboard section of the Elm Vega-Lite Walkthrough for more details.
Interactivity
Interaction is enabled by creating selections that may be combined with the kinds of specifications already described. Selections involve three components:
- Events are those actions that trigger the interaction such as clicking at a location on screen or pressing a key.
- Points of interest are the elements of the visualization with which the interaction occurs, such as the set of points selected on a scatterplot.
- Predicates (i.e. Boolean functions) identify whether or not something is included in the selection. These need not be limited to only those parts of the visualization directly selected through interaction.
Arguments
| :: Text | The selection name |
| -> Text | The title for the plot |
| -> [PropertySpec] |
The next several plots show different types of selection - select a single point, a range of plots, or follow the mouse - and all have the same basic structure. To avoid repetition, and mistakes, I am going to introduce a helper function which creates the plot structure but without the selection definition, and then use that to build up the plots.
The helper function, selectionProperties, takes two arguments, which are
the selection name and the plot title. The selection name is used
to identify the selection, as a visualization can support multiple
selections, and the plot title has been added mainly to show some
minor customization (the use of TOrient to move the title to the
bottom).
The definition of this helper function is:
selectionProps selName label =
let posOpts field = [ PName field
, PmType Quantitative
, PScale [ SZero False ]
]
enc = encoding
. position X (posOpts "Gmag")
. position Y (posOpts "plx")
. color [ MSelectionCondition (SelectionName selName)
[ MName "Cluster", MmType Nominal ]
[ MString "grey" ]
]
. opacity [ MSelectionCondition (SelectionName selName)
[ MNumber 1.0 ]
[ MNumber 0.3 ]
]
. size [ MSelectionCondition (SelectionName selName)
[ MNumber 40 ]
[ MNumber 5 ]
]
trans = transform
. filter (FExpr "datum.DE_ICRS < -20")
in [ gaiaData
, trans []
, mark Point []
, enc []
, title label [ TOrient SBottom ]
]
The three non-selection-related features added here are that
SZero is used to tell Vega Lite that we do not need 0 displayed
on either axis, which leads to a "tight" bounding box around
the data, a filter is used to select a subset of rows, namely
only those with a declination less than -20 (via FExpr),
and the plot title is moved to the bottom with TOrient.
The main change is that the selection is used in the encoding section,
identified by name, using SelectionName and the supplied
argument. It is used as a filter for the encoding section, where
MSelectionCondition defines the properties to use
when the selection occurs (the first list of properties)
and when it does not (the second list). This is used for
three different encodings:
color, where the selected star is labelled by its cluster color, and all the other are grey;opacity, so that the selected star is fully opaque whereas un-selected stars are partially transparent;- and
size, so that the selected star is much bigger than the others.
When no selection has been made - such as when the visualization is first created - then all points are encoded with the "selected" case (so colorful, fully opaque, and large in this case).
singleSelection :: VegaLite Source #
The actual plot just requires the selection information to be defined and then added to the plot properties:
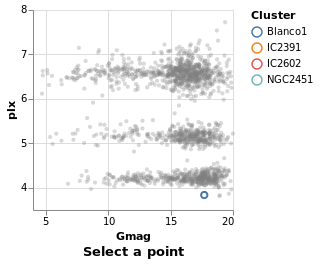
Open this visualization in the Vega Editor
let selLabel = "picked"
sel = selection
. select selLabel Single []
in toVegaLite (sel [] : selectionProperties selLabel "Select a point")
The selection function is used to define the selection, via one or
more applications of the select function. The form of select is
that the selection is named, in this case we use "picked", and the
type is given (a Single click), and then options, which in our case
there aren't any, so an empty list is used.
Note that hvega does not track the selection names, and will allow
you to use a name that you have not defined.
nearestSelection :: VegaLite Source #
The only change here is to add a property to the selection - that
is Nearest True
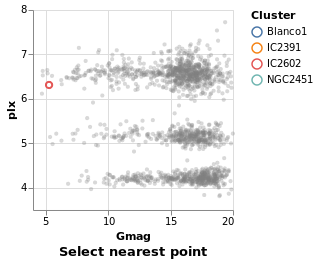
Open this visualization in the Vega Editor
let selLabel = "picked"
sel = selection
. select selLabel Single [ Nearest True ]
in toVegaLite (sel [] : selectionProperties selLabel "Select nearest point")
One consequence of this change is that once a point has been selected you
can not remove this (i.e. un-select the point). This is in contrast to
singleSelection, where clicking on an area with no stars would remove the
previous selection. The Clear property can be added to the list
to define a way to clear the selection.
multiSelection :: VegaLite Source #
The selection can easily be changed to allow multiple stars
to be selected, using shift-click, by swapping from Single to Multi.

Open this visualization in the Vega Editor
let selLabel = "this is just a label"
sel = selection
. select selLabel Multi []
in toVegaLite (sel [] : selectionProperties selLabel "Shift click to select points")
eventSelection :: VegaLite Source #
We can take advantage of browser event by using On to define which
event to use, such as mouse movement over points:
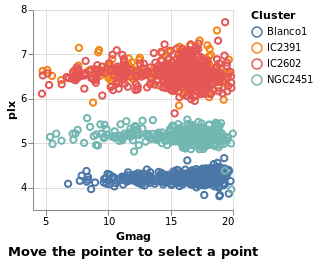
Open this visualization in the Vega Editor
let selLabel = "picked"
sel = selection
. select selLabel Multi [ On "mouseover" ]
in toVegaLite (sel [] : selectionProperties selLabel "Move the pointer to select a point")
The supported list of events is described in the Vega Event-Stream Selectors documentation.
The addition of Nearest True to the list of properties
sent to select would avoid the flickering, as the mouse moves
between the stars.
intervalSelection :: VegaLite Source #
The final Selection value is Interval,
which lets you drag a rectangle to select the interior points:
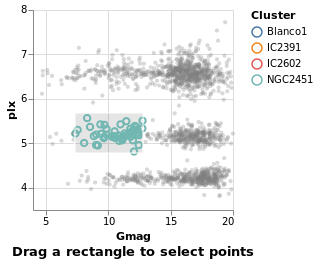
Open this visualization in the Vega Editor
let selLabel = "naming is hard"
sel = selection
. select selLabel Interval [ ]
in toVegaLite (sel [] : selectionProperties selLabel "Drag a rectangle to select points")
intervalSelectionY :: VegaLite Source #
The default interval option is to select a rectangle, but it can be restricted -
such as to select all items within a range along a given axis
using Encodings:
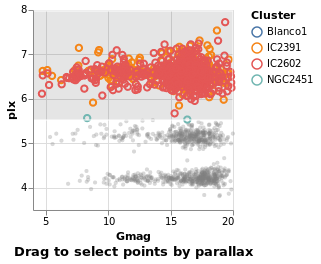
Open this visualization in the Vega Editor
let selLabel = "naming is still hard"
sel = selection
. select selLabel Interval [ Encodings [ ChY ] ]
in toVegaLite (sel [] : selectionProperties selLabel "Drag to select points by parallax")
We'll come back to further things to do with interval selections
when we get to interactive plots below (see bindScales).
Selection Transformations
Simple selections as described above create sets of selected data marks based directly on what was interacted with by the user. Selection transformations allow us to project that direct selection onto other parts of our dataset.
transformSelection :: VegaLite Source #
For example, we can adjust the visualization to select all stars in the same cluster, which is useful in this case since the Blanco1 and IC2391 clusters occupy the same space in the magnitude-parallax plane.
This is invoked simply by adding the Fields constructor to the select
parameters naming the fields onto which we wish to project our selection.
Additionally, we have set the default selection with Empty so that if
nothing is selected, the selection is empty
(as we have previously seen, without this the default selection is the
entire encoded dataset).
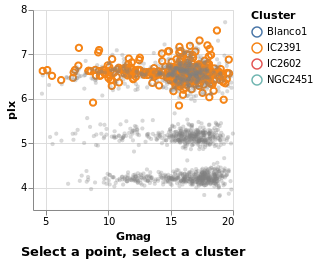
Open this visualization in the Vega Editor
let sel = selection
. select "pick" Single [ Fields [ "Cluster" ]
, Empty
, Nearest True
]
in toVegaLite (sel [] : selectionProperties "pick" "Select a point, select a cluster")
Selection and bindings
Selection need not be limited to direct interaction with the visualization marks. We can also bind the selection to other user-interface components.
widgetSelection :: VegaLite Source #
The Elm Vega-Lite walkthrough uses a dataset which has a
column for which a range-slider makes sense. The dataset I'm
using is less rich, and so I am going to use a HTML select
widget - a drop-down list of values - instead. This lets
the user select all stars from a given cluster, and is
introduced with the Bind and ISelect constructors.
The InOptions list is given the values of the Cluster column that
can be selected: I start with a value not in the list (none) just to
indicate that no values are selected, and then the list of clusters in
this sub-sample (remembering that selectionProperties applies a
declination cut off). Eagle-eyed readers will note that the cluster
names in this list (the clusters variable) end in spaces: this is
because the input data file has the cluster names stored in an
eight-character field, even though it is a tab-separated file.
This surprised me when I first tried this visualization, and
using the value "Blanco1" did not select anything! Isn't working
with data so much fun!
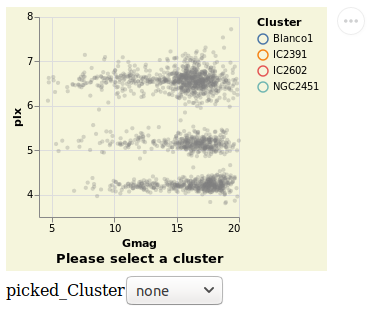
Open this visualization in the Vega Editor
let picked = "picked"
clusters = [ "none", "Blanco1 ", "IC2391 ", "IC2602 ", "NGC2451 " ]
sel = selection
. select picked Single [ Fields [ "Cluster" ]
, Bind [ ISelect "Cluster" [ InOptions clusters ] ]
, Empty
]
conf = configure
. configuration (Background "beige")
in toVegaLite (conf [] :
sel [] :
selectionProperties picked "Please select a cluster")
Note that the selection works both ways: the HTML widget can be used to select a cluster, and clicking on a point on the visualization will update the HTML widget.
Unlike the other plots shown in the tutorial, this is a screen grab
rather than a PNG file created by Vega Embed. The background color was
changed - following the approach used in
stripPlotWithBackground - to show where the visualization "ends" and
the HTML select element starts. It also shows the Vega Embed "drop-down"
menu in the top-right corner, namely the three dots in a circle.
bindScales :: VegaLite Source #
The selection can also be bound to an axis (or both axes, as in this
case), using BindScales (applying it to the intervalSelectionY plot).
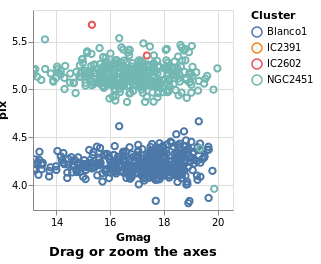
Open this visualization in the Vega Editor
let picked = "picked"
sel = selection
. select picked Interval [ Encodings [ ChX, ChY ], BindScales ]
in toVegaLite (sel [] : selectionProperties picked "Drag or zoom the axes")
The image here was created after panning and zooming into the data.
Multiple Coordinated Views
One of the more powerful aspects of selection-based interaction is in coordinating different views – a selection of a data subset is projected onto all other views of the same data.
coordinatedViews :: VegaLite Source #
The following plot doesn't contain anything new, but allows us to
select a rectangular-range on one plot, and see the same selection
automatically applied to the other plots. This is achieved by
combining the repeat fuction with the selection; this causes the
selection to be projected across all views as it is duplicated.
For this example we use all the clusters, rather than the subset of Southern ones. One trick I use is to convert the Right Ascension values (which have a domain of 0 to 360 degrees, and wrap around at the 0/360 mark), into their cosine values (remembering to convert to radians first), and display that instead. This ensures the "Blanco1" cluster members are spatially co-located on this axis - with values close to 1 - rather than appearing near 0 and 360.
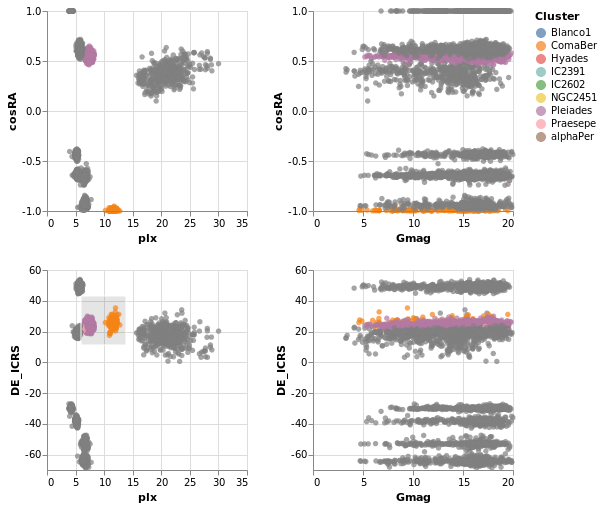
Open this visualization in the Vega Editor
let enc = encoding
. position X [ PRepeat Column, PmType Quantitative ]
. position Y [ PRepeat Row, PmType Quantitative ]
. color
[ MSelectionCondition (SelectionName "picked")
[ MName "Cluster", MmType Nominal ]
[ MString "grey" ]
]
sel = selection
. select "picked" Interval [ ]
trans = transform
. calculateAs "cos(datum.RA_ICRS * PI / 180)" "cosRA"
spec = asSpec
[ gaiaData
, trans []
, mark Circle []
, enc []
, sel []
]
in toVegaLite
[ repeat
[ RowFields [ "cosRA", "DE_ICRS" ]
, ColumnFields [ "plx", "Gmag" ]
]
, specification spec
]
coordinatedViews2 :: VegaLite Source #
If the interval selection is bound the the axes with BindScales
then we can zoom and pan the related plots - i.e. changing the
range displayed in one plot will also change the two plots that
it shares an axis with in this two by two arrangement. The conditional
encoding of the color channel has also been removed.
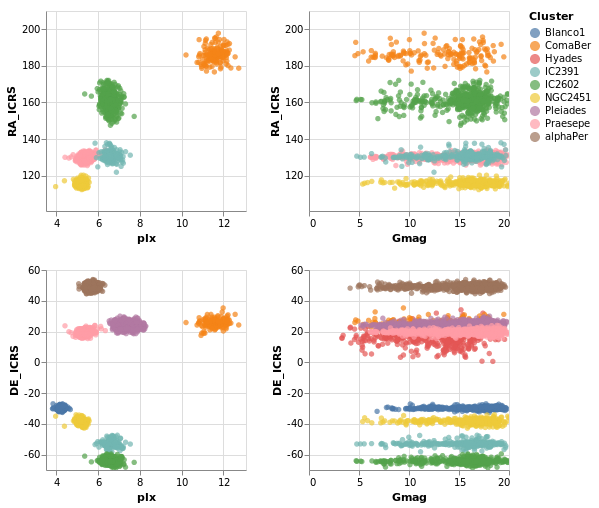
Open this visualization in the Vega Editor
The image was captured after panning and zooming in the "parallax-RA_ICRS" plot.
let enc = encoding
. position X [ PRepeat Column, PmType Quantitative ]
. position Y [ PRepeat Row, PmType Quantitative ]
. color [ MName "Cluster", MmType Nominal ]
sel = selection
. select "picked" Interval [ BindScales ]
spec = asSpec
[ gaiaData
, mark Circle []
, enc []
, sel []
]
in toVegaLite
[ repeat
[ RowFields [ "RA_ICRS", "DE_ICRS" ]
, ColumnFields [ "plx", "Gmag" ]
]
, specification spec
]
The "cosine" transformation has been removed in comparison to
coordinatedViews.
contextAndFocus :: VegaLite Source #
The ability to determine the scale of a chart based on a selection is useful in implementing a common visualization design pattern, that of 'context and focus' (or sometimes referred to as 'overview and detail on demand'). We can achieve this by setting the scale of one view based on the selection in another. The detail view is updated whenever the selected region is changed through interaction:

Open this visualization in the Vega Editor
let sel = selection . select "brush" Interval [ Encodings [ ChY ] ]
encContext = encoding
. position X [ PName "Gmag", PmType Quantitative, PScale [ SZero False ] ]
. position Y [ PName "plx", PmType Quantitative ]
specContext = asSpec [ width 400
, height 80
, sel []
, mark Point []
, encContext []
, title "Select a Y range to zoom in below" []
]
encDetail = encoding
. position X [ PName "Gmag"
, PmType Quantitative
, PScale [ SZero False ]
, PAxis [ AxNoTitle ]
]
. position Y [ PName "plx"
, PmType Quantitative
, PScale [ SDomain (DSelection "brush") ]
]
. color [ MName "Cluster", MmType Nominal ]
specDetail =
asSpec [ width 400, mark Point [], encDetail [] ]
in toVegaLite
[ gaiaData
, vConcat [ specContext, specDetail ]
]
Not shown here, but selecting a range of y-values in the top plot
(specContext) will cause the second plot (specDetail) to zoom
in on that range, as the selection is bound to the y axis of this
plot via DSelection.
Cross-filtering
The final example in this section brings together ideas of view composition and interactive selection with data filtering by implementing cross-filtering: the selection of a subset of the data in one view then only displaying that data in the other views.
crossFilter :: VegaLite Source #
Here we show distributions of the four main numeric quantities
in the dataset - position, magnitude, and prallax - using the
totalEnc encoding, and add a second layer which repeats this
data but with a different color (selectedEnc), and that is tied to
the interval-selection along the x axis (ChX).
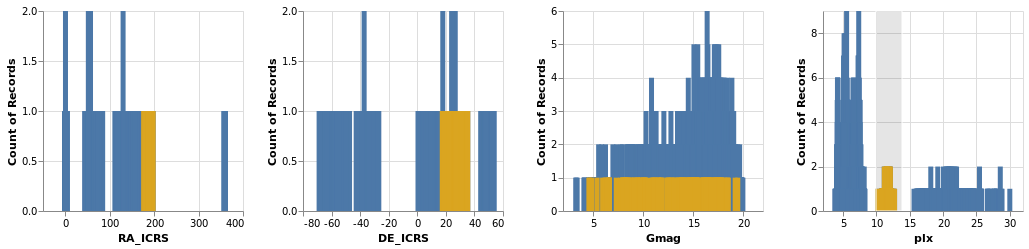
Open this visualization in the Vega Editor
Selecting a small range of parallax values in the fourth plot highlights the associated data in the other three plots.
let sel = selection . select "brush" Interval [ Encodings [ ChX ] ]
filterTrans = transform . filter (FSelection "brush")
-- borrow a function from Elm
pQuant = PmType Quantitative
totalEnc = encoding
. position X [ PRepeat Column, pQuant ]
. position Y [ PAggregate Count, pQuant ]
selectedEnc = totalEnc
. color [ MString "goldenrod" ]
in toVegaLite
[ repeat [ ColumnFields [ "RA_ICRS", "DE_ICRS", "Gmag", "plx" ] ]
, specification $
asSpec
[ gaiaData
, layer
[ asSpec [ mark Bar [], totalEnc [] ]
, asSpec [ sel [], filterTrans [], mark Bar [], selectedEnc [] ]
]
]
]
Errors: lines, bars, bands, and boxes
Here we dive into some of the ways for representing the spread of a value, focussing on the "error" of a variable.
errorManual :: VegaLite Source #
We have already seen "error bars" in the layeredDiversion plot, where
the Rule type was used to draw a line between the Y and Y2
encodings. In that example the two positions were calculated "on the
fly" by Vega-Lite (using the Min and Max aggregation operations).
In this example I use the data to calculate the display range,
namely plx - e_plx to plx + e_plx. These are mapped to the
X and X2 channels (not because it makes a better visualization,
but just to show you can create lines along the x axis), and a
small-multiples approach is used to separate out the clusters, but
only after a filter designed to select the two clusters - with the
"most interesting" data for this plot - has been applied.

Open this visualization in the Vega Editor
let trans = transform
. filter (FExpr "datum.Cluster[0] == 'C' || datum.Cluster[0] == 'H'")
. calculateAs "datum.plx - datum.e_plx" "plx_lo"
. calculateAs "datum.plx + datum.e_plx" "plx_hi"
errorEnc = encoding
. position X [ PName "plx_lo"
, PmType Quantitative
, PScale [SZero False]
, PAxis [ AxTitle "parallax (mas)" ]
]
. position X2 [ PName "plx_hi" ]
. position Y [ PName "Gmag", PmType Quantitative ]
. column [ FName "Cluster", FmType Nominal ]
sel = selection
. select "picked" Interval [ BindScales ]
in toVegaLite [ gaiaData
, trans []
, errorEnc []
, mark Rule []
, sel []
]
For the interested reader, it was the calculation of the "plx_hi"
column that lead me to the discovery that the columns were being
read in as a string, and the introduction of the Parse option
to gaiaData.
As can be seen, the e_plx terms are generally very small. This is
good for anyone using the data, as we want precise measurements, but
makes it harder for me to come up with meaningful visualizations! I
have taken advantage of the BindScales interaction to zoom in on
a subset of stars which show larger parallax errors:

errorAuto :: VegaLite Source #
Alternatively, I could have made life simpler for myself and used
the ErrorBar (or ErrorBand) mark type, together with XError
(or YError) to indicate that the channel gives the offset from
the central value. For this visualization I restrict to a single
cluster (since I now know there's only one in this sample which
begins with C), but retain the column encoding as a means
to getting a useful title. I've also switched things so that the
errors are back along the y axis.
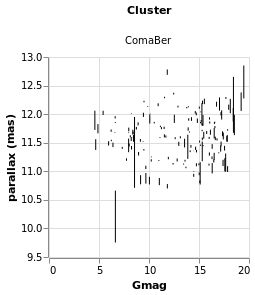
Open this visualization in the Vega Editor
let trans = transform
. filter (FExpr "datum.Cluster[0] == 'C'")
errorEnc = encoding
. position Y [ PName "plx"
, PmType Quantitative
, PScale [SZero False]
, PAxis [ AxTitle "parallax (mas)" ]
]
. position YError [ PName "e_plx" ]
. position X [ PName "Gmag", PmType Quantitative ]
. column [ FName "Cluster", FmType Nominal ]
in toVegaLite [ gaiaData
, trans []
, errorEnc []
, mark ErrorBar []
]
errorBars :: VegaLite Source #
In this plot the error range is calculated by Vega-Lite,
and is taken from the standard deviation of the Gmag field
(StdDev). The MTicks and MRule constructors are used to
color the different parts of the error bars. Since the error bar
does not reference the central value, a separate layer is used
to add a square symbol (SymSquare) at the average (Mean)
value of the distribution.
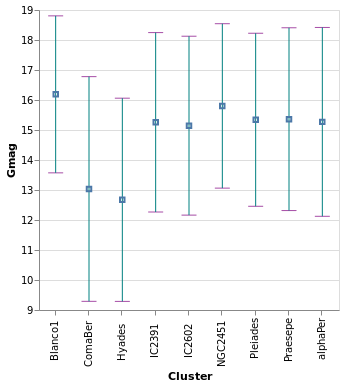
Open this visualization in the Vega Editor
let cluster = position X [ PName "Cluster", PmType Nominal ]
barOpts = [ MExtent StdDev
, MTicks [ MColor "purple" ]
, MRule [ MColor "teal" ]
]
range = [ mark ErrorBar barOpts
, encoding
. position Y [ PName "Gmag"
, PmType Quantitative
, PScale [ SZero False ]
]
$ []
]
center = [ mark Point [ MShape SymSquare, MSize 20 ]
, encoding
. position Y [ PName "Gmag"
, PmType Quantitative
, PmType Mean
]
$ []
]
in toVegaLite
[ gaiaData
, encoding (cluster [])
, layer (map asSpec [ range, center ])
, width 300
, height 300
]
errorBand :: VegaLite Source #
The next plot shows the ErrorBand mark, which fills the area between
the chosen range with a color, and optional borders. Here the
blue band shows the calculated standard deviation - as used in
errorBars - and the gray band with borders shows the inter-quartile
range. On top of these are drawn the median (blue) and median (green
dashed) lines.

Open this visualization in the Vega Editor
let posY extra = position Y ([ PName "Gmag"
, PmType Quantitative
, PScale [ SZero False ]
] ++ extra) []
bands = [ [ encoding (posY [])
, mark ErrorBand [ MExtent StdDev ]
]
, [ encoding (posY [])
, mark ErrorBand [ MExtent Iqr
, MBorders [ MStrokeDash [ 6, 2 ] ]
, MColor "gray"
]
]
, [ encoding (posY [ PAggregate Median ])
, mark Line []
]
, [ encoding (posY [ PAggregate Mean ])
, mark Line [ MColor "green"
, MStrokeDash [ 6, 2, 4, 2 ]
]
]
]
in toVegaLite
[ gaiaData
, encoding (position X [ PName "Cluster", PmType Nominal ] [])
, layer (map asSpec bands)
, width 300
, height 300
, title "Comparing ranges" []
]
Note that I don't think this is a good visualization
for this particular dataset, since it implies there's a
connection or correlation between clusters, as given by the
x-axis ordering, but the aim here is to show how to use hvega
rather than creating sensible plots!
An alternative visualization of a distribution is the "box and
whiskers" plot, which can be achieved in hvega with the
Boxplot mark. The example below shows the default settings, but
the various components can be controlled with MBox, MMedian,
MOutliers, and MTicks.
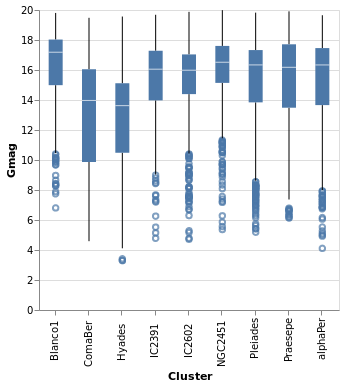
Open this visualization in the Vega Editor
toVegaLite
[ gaiaData
, encoding
. position X [ PName "Cluster", PmType Nominal ]
. position Y [ PName "Gmag", PmType Quantitative ]
$ []
, mark Boxplot []
, width 300
, height 300
]
The Boxplot option supports two different "ranges":
- the default is the Tukey Box plot, where the whiskers span
a range
Q1 - k * IQRtoQ3 + k * IQR,IQR = Q3 - Q1,Q1andQ3are the lower and upper inter-quartile values (so 25 and 75 per cent of the distribution), andkdefaults to 1.5 but can be changed withIqrScale; - or
ExRange, which uses the full range of the data (i.e. minimum to maximum values).
comparingErrors :: VegaLite Source #
Here I combine errorBox with smallMultiples2 so we can compare
the distribution (from the histogram) with that from the box plot.
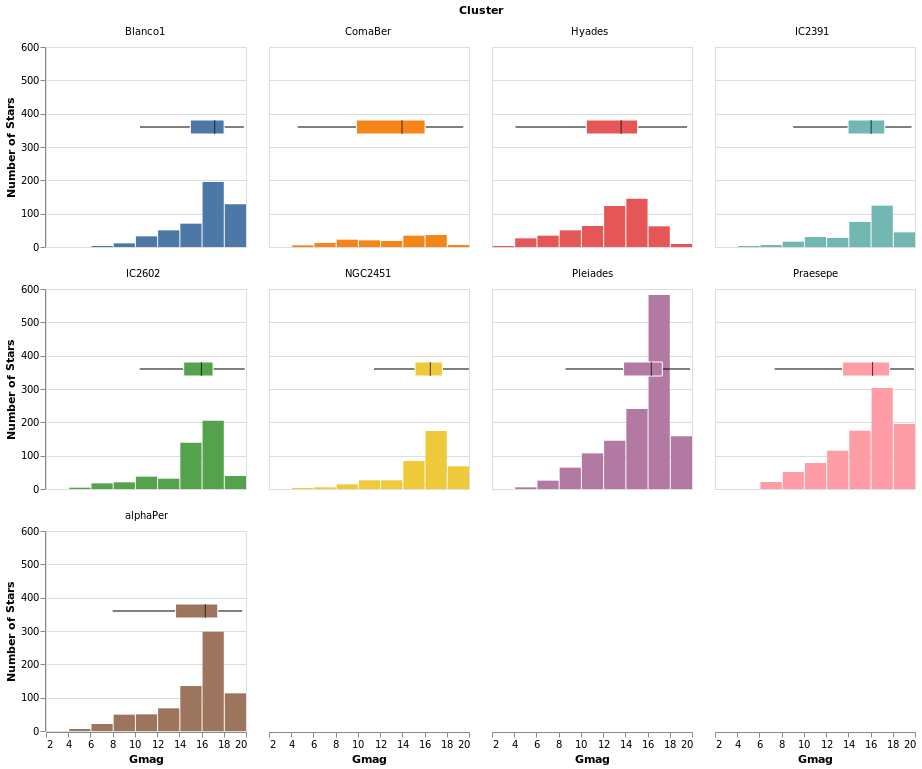
Open this visualization in the Vega Editor
let histEnc = encoding
. position X [ PName "Gmag", PmType Quantitative, PBin [] ]
. position Y yAxis
. color [ MName "Cluster", MmType Nominal, MLegend [] ]
errEnc = encoding
. position X [ PName "Gmag", PmType Quantitative ]
. position Y [ PNumber 80 ]
. color [ MName "Cluster", MmType Nominal, MLegend [] ]
yAxis = [ PAggregate Count
, PmType Quantitative
, PAxis [ AxTitle "Number of Stars" ]
]
boxOpts = [ MMedian [ MColor "black" ]
, MBox [ MStroke "white" ]
, MNoOutliers
]
histSpec = asSpec [ mark Bar [], histEnc [] ]
errSpec = asSpec [ mark Boxplot boxOpts, errEnc [] ]
combinedSpec = asSpec [ layer [ histSpec, errSpec ] ]
in toVegaLite
[ gaiaData
, columns 4
, facetFlow [ FName "Cluster", FmType Nominal ]
, specification combinedSpec
]
The main additions here are the configuration of the box plot - with
MMedian, MBox (used to ensure the box is visually distinct from
the bar for the Pleiades cluster, where they overlap), and MNoOutliers
(to turn off the display of the outliers) - and the use of PNumber
to define the location on the y axis of the boxplot visualization.
Note that PNumber is defined in pixel units, with 0 being the
top of the visualization and 80 was found by trial and error.
Dashboard-esque
In the following visualization I try to combine as many of the concepts we have explored in this tutorial into one. There are layers, combined visualizations, and a selection that ties the different plots together.
combinedPlot :: VegaLite Source #
This is based on the Marginal Histogram example from the Vega-Lite Example Gallery. There is very-little new in this plot, in that pretty-much everything has been shown before. However, there are some interesting wrinkles, such as
- combining multiple plots, in this case the "top" area - which is
a histogram on top of a plot which is itself a "map" and
a histogram - and "bottom" area - which is just a point plot -
requires judicious use of
asSpec; - selection works in both the main plots - the "map" and "point"
plots - to highlight all stars in the same cluster, and I was
pleasantly surprised to find out I could just use the same selection
specification (
selCluster) in both (hopefully I am not just ejnoying a hole in the Vega Lite specification); - I have been perhaps too defensive in defining the Right Ascension
and Declination axes in the relevant plots, as I want to make sure
the histogram bins and plot axes are well aligned (that is the
NiceFalse - I am not 100% sure I understand what is going on with the
grid labels on the Declination axis, as I had thought I was
asking for marks every 15 degrees, but the plot shows them
every 30 degrees (however, if I change the
deTicksarray then the marks change in ways I currently do not understand); - and I have decided to display Right Ascension in hours, rather than degrees, because why have one way to measure a value when you can have many!
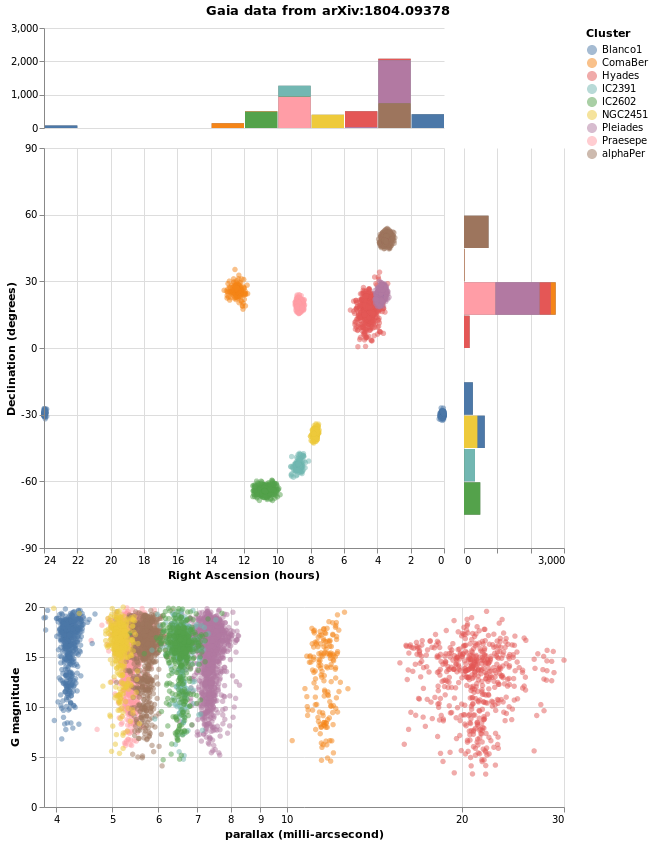
let trans = transform
. calculateAs "datum.RA_ICRS / 15" "RA"
quant n = [ PName n, PmType Quantitative ]
big = 400
small = 100
wmain = width big
hmain = height big
wsub = width small
hsub = height small
noTitle = PAxis [ AxNoTitle ]
raAxis = [ PScale [ SDomain (DNumbers [ 0, 24 ])
, SNice (IsNice False)
]
, PSort [ Descending ]
, PAxis [ AxTitle "Right Ascension (hours)" ]
]
deMin = -90
deMax = 90
deStep = 15
-- we do not get ticks/grids at all these values, but it does
-- something (e.g. if do not specify the axis ticks are different)
--
deTicks = Numbers [ deMin, deMin + deStep .. deMax ]
deAxis = [ PScale [ SDomain (DNumbers [ deMin, deMax ])
, SNice (IsNice False)
]
, PAxis [ AxTitle "Declination (degrees)"
, AxValues deTicks
]
]
colorEnc = color [ MSelectionCondition (SelectionName "pick")
[ MName "Cluster", MmType Nominal ]
[ MString "grey" ]
]
mapEnc = encoding
. position X (quant "RA" ++ raAxis)
. position Y (quant "DE_ICRS" ++ deAxis)
. colorEnc
circleMark = mark Circle [ MOpacity 0.5 ]
mapSpec = asSpec [ mapEnc []
, circleMark
, wmain
, hmain
, selCluster []
]
-- histogram of the RA values
--
raBinning = [ PBin [ Extent 0 24
, Step 2
, Nice False
]
, PSort [ Descending ]
, PAxis []
]
-- histogram of the Declination values
--
deBinning = [ PBin [ Extent deMin deMax
, Step deStep
, Nice False
]
, PAxis []
]
histAxis = [ PAggregate Count
, PmType Quantitative
, noTitle
, PScale [ SDomain (DNumbers [ 0, 3000 ]) ]
]
raEnc = encoding
. position X (quant "RA" ++ raBinning)
. position Y histAxis
deEnc = encoding
. position Y (quant "DE_ICRS" ++ deBinning)
. position X histAxis
allRA = [ raEnc []
, mark Bar [ MColor "gray" ]
]
filtRA = [ filterCluster []
, raEnc
. colorEnc
$ []
, mark Bar []
]
allDE = [ deEnc []
, mark Bar [ MColor "gray" ]
]
filtDE = [ filterCluster []
, deEnc
. colorEnc
$ []
, mark Bar []
]
raSpec = asSpec [ wmain, hsub, layer [ asSpec allRA, asSpec filtRA ] ]
deSpec = asSpec [ hmain, wsub, layer [ asSpec allDE, asSpec filtDE ] ]
borderSpacing = 20
mapAndDecSpec = asSpec [ spacing borderSpacing
, bounds Flush
, hConcat [ mapSpec, deSpec ]
]
histSpecs = [ raSpec, mapAndDecSpec ]
-- select the cluster which the star belongs to; do not use
-- "nearest click" as that means a user can not cancel the
-- selection.
--
pick = "pick"
selCluster = selection
. select pick Single [ Fields [ "Cluster" ] ]
filterCluster = transform
. filter (FSelection pick)
plxOpts = [ PScale [ SType ScLog, SNice (IsNice False) ]
, PAxis [ AxTitle "parallax (milli-arcsecond)" ]
]
gmagOpts = [ PAxis [ AxTitle "G magnitude" ] ]
encData = encoding
. position X (quant "plx" ++ plxOpts)
. position Y (quant "Gmag" ++ gmagOpts)
parallaxSpec = asSpec [ width (big + borderSpacing + small)
, encData
. colorEnc
$ []
, circleMark
, selCluster []
]
allSpecs = [ asSpec [ spacing borderSpacing
, bounds Flush
, vConcat histSpecs
]
, parallaxSpec ]
in toVegaLite
[ gaiaData
, trans []
, vConcat allSpecs
-- remove the "other" axis (e.g. top of Y, right for X)
, configure
. configuration (View [ ViewStroke (Just "transparent") ])
$ []
, title "Gaia data from arXiv:1804.09378" [ TAnchor AMiddle ]
]
Here is the visualization after selecting a star:
